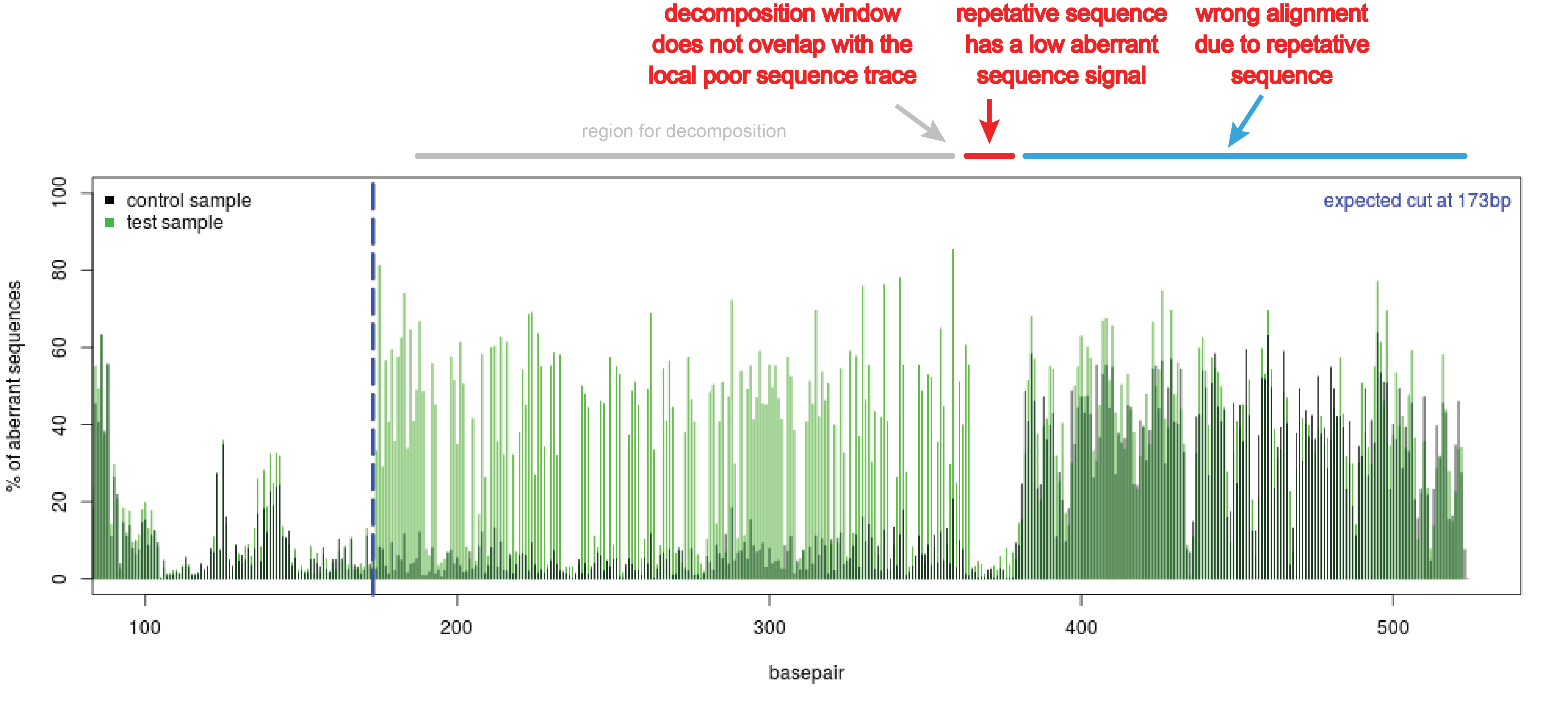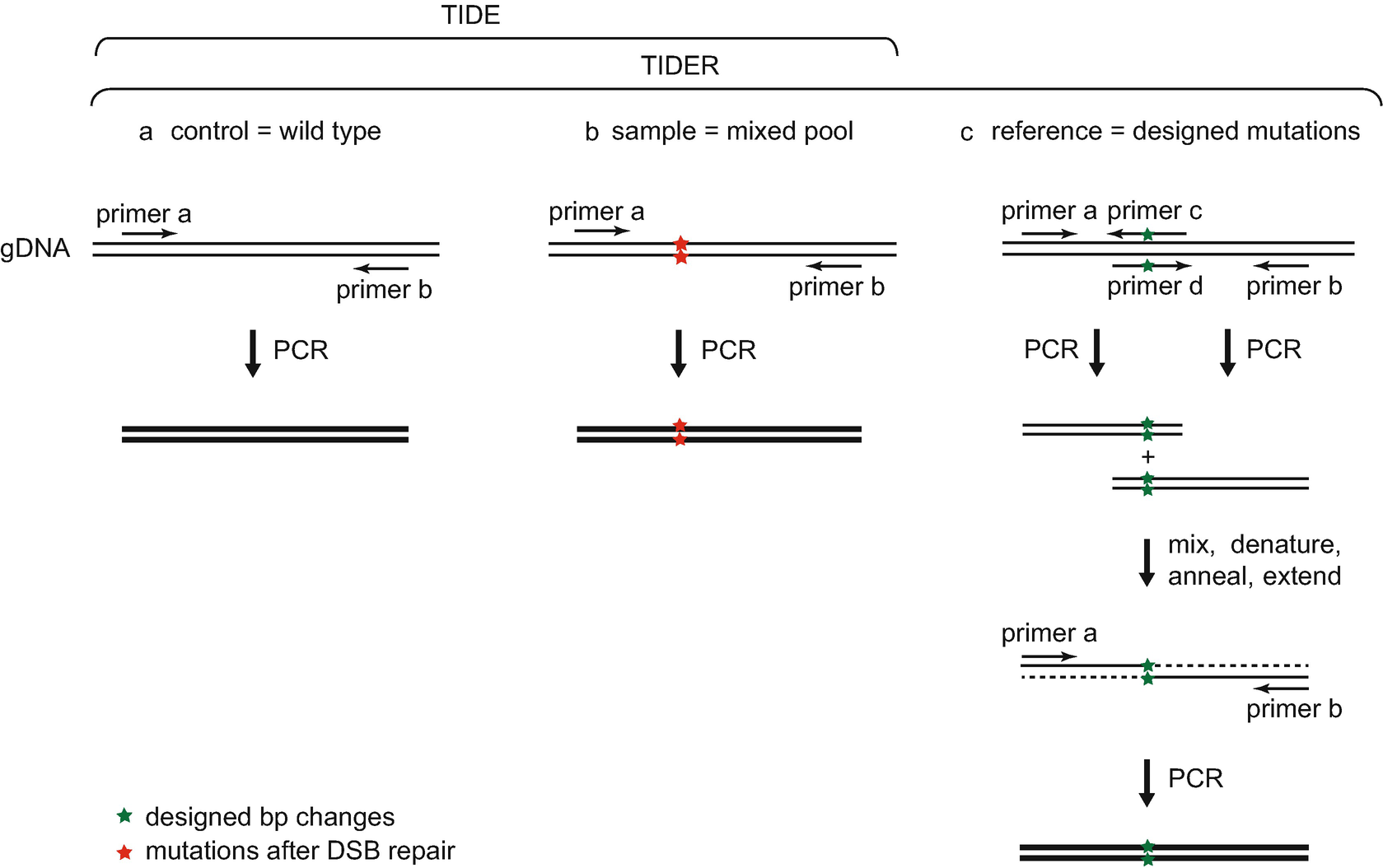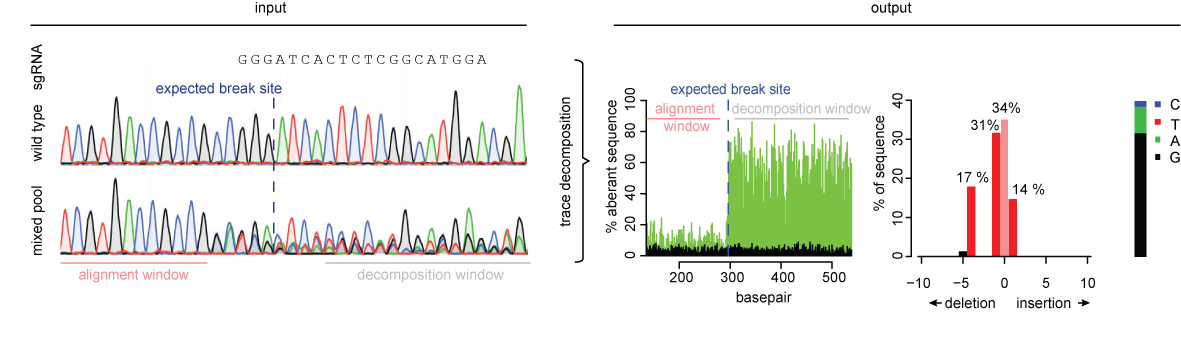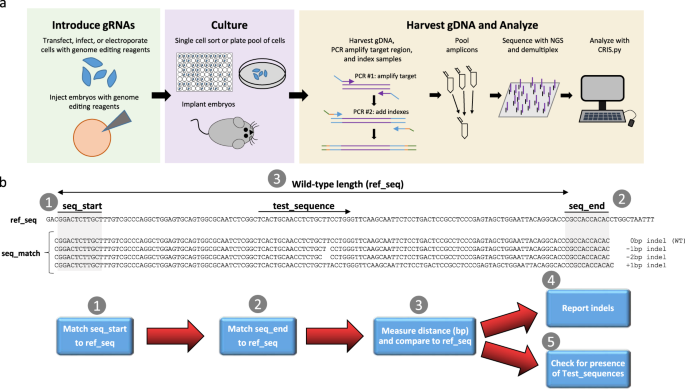
CRIS.py: A Versatile and High-throughput Analysis Program for CRISPR-based Genome Editing | Scientific Reports
![Cells | Free Full-Text | A Versatile Strategy to Reduce UGA-Selenocysteine Recoding Efficiency of the Ribosome Using CRISPR-Cas9-Viral-Like-Particles Targeting Selenocysteine-tRNA[Ser]Sec Gene | HTML Cells | Free Full-Text | A Versatile Strategy to Reduce UGA-Selenocysteine Recoding Efficiency of the Ribosome Using CRISPR-Cas9-Viral-Like-Particles Targeting Selenocysteine-tRNA[Ser]Sec Gene | HTML](https://www.mdpi.com/cells/cells-08-00574/article_deploy/html/images/cells-08-00574-g004.png)
Cells | Free Full-Text | A Versatile Strategy to Reduce UGA-Selenocysteine Recoding Efficiency of the Ribosome Using CRISPR-Cas9-Viral-Like-Particles Targeting Selenocysteine-tRNA[Ser]Sec Gene | HTML

Increasing CRISPR Efficiency and Measuring Its Specificity in HSPCs Using a Clinically Relevant System: Molecular Therapy - Methods & Clinical Development

Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases - ScienceDirect
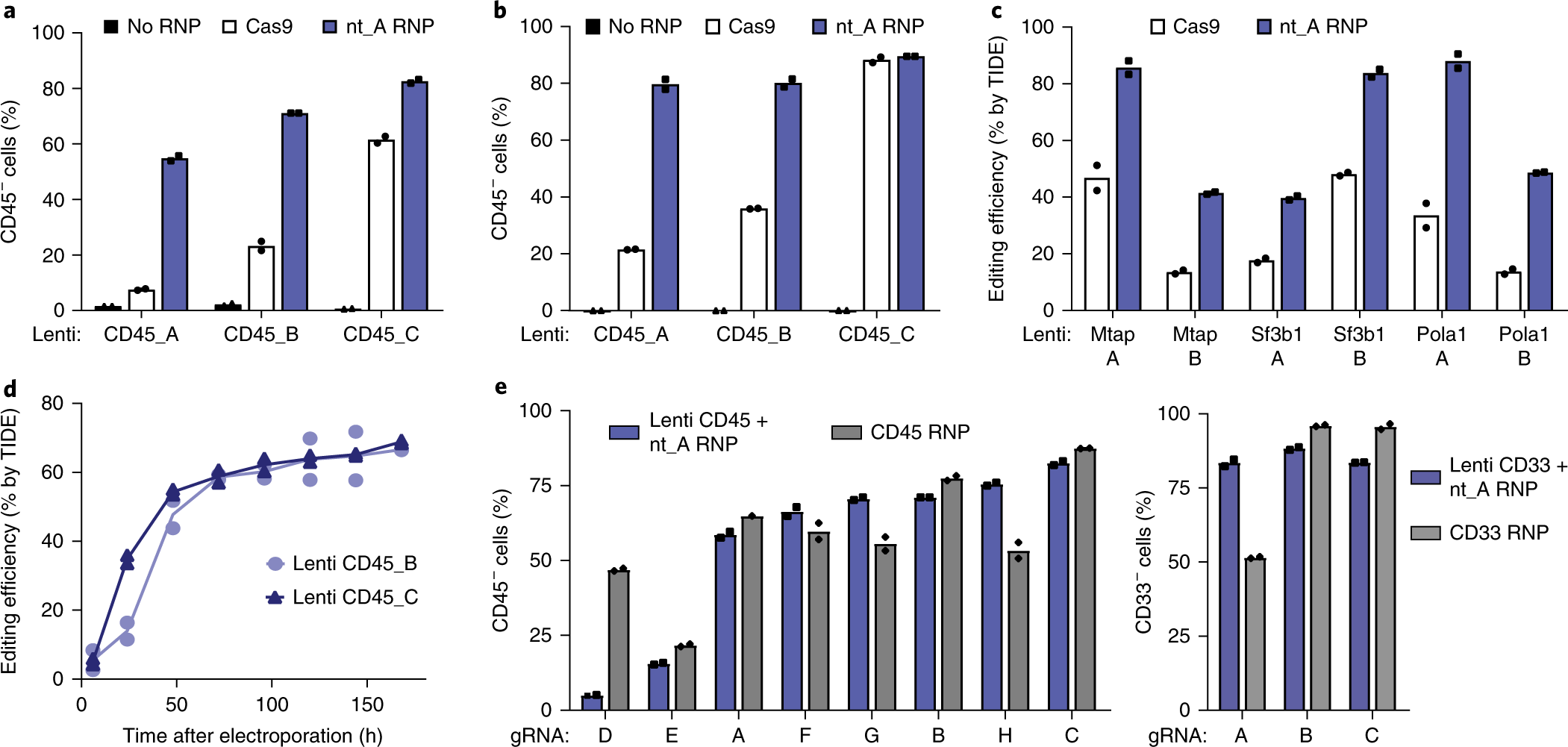
Guide Swap enables genome-scale pooled CRISPR–Cas9 screening in human primary cells | Nature Methods
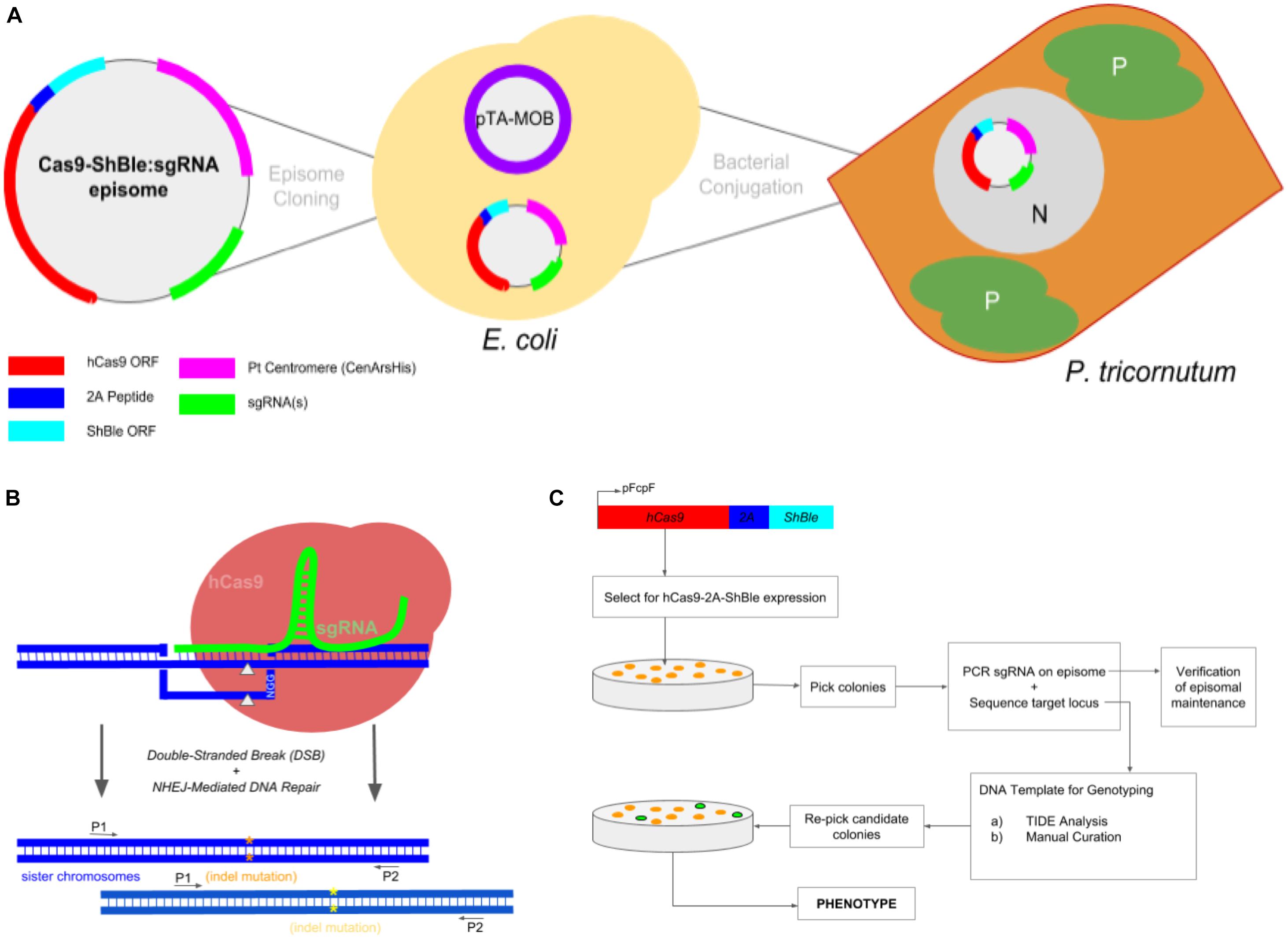
Frontiers | Multiplexed Knockouts in the Model Diatom Phaeodactylum by Episomal Delivery of a Selectable Cas9 | Microbiology

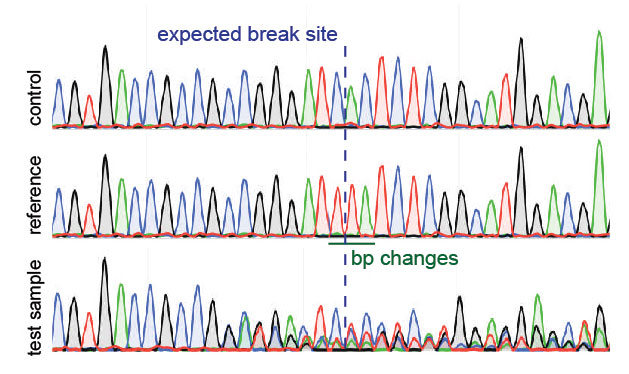
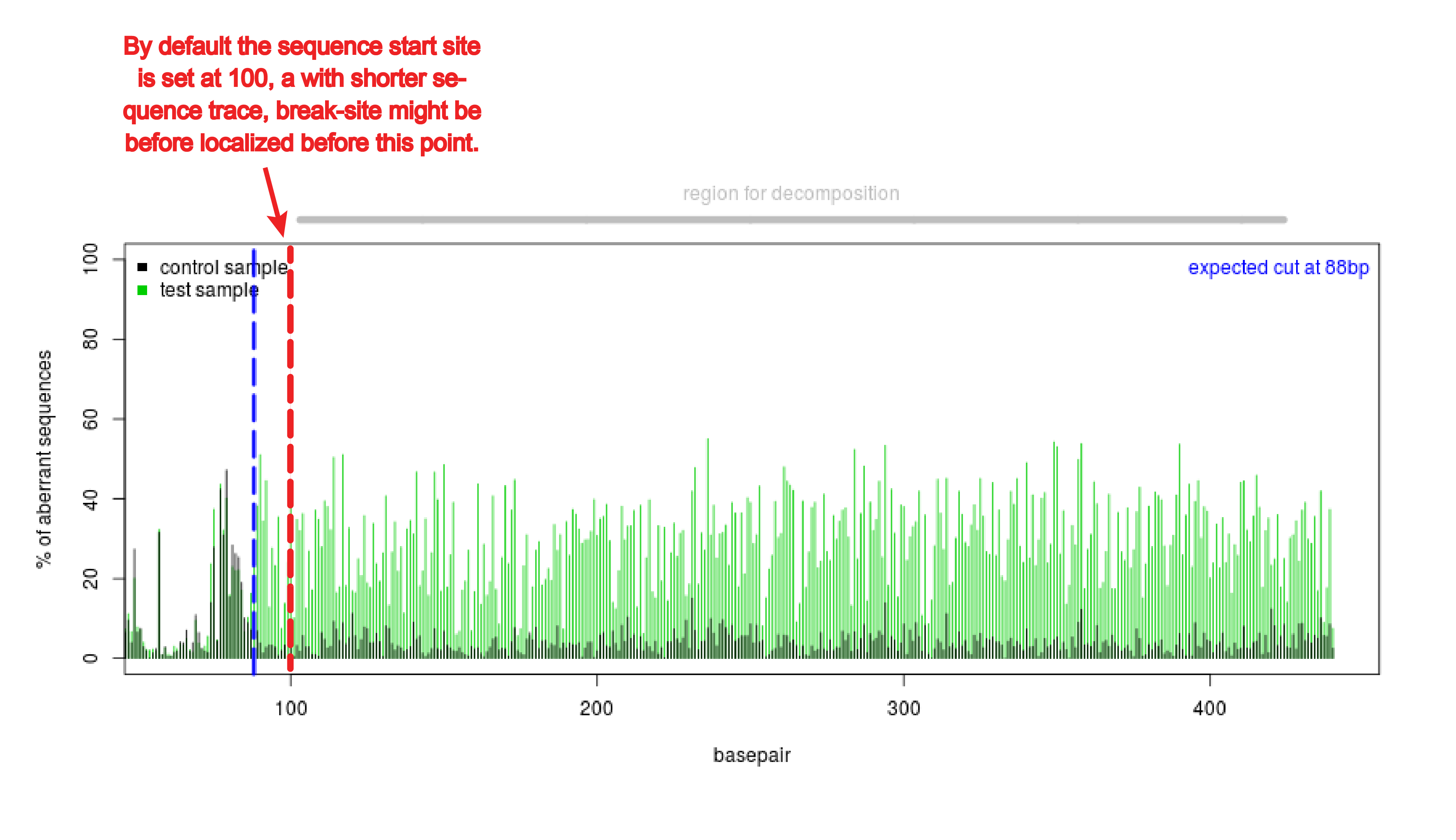
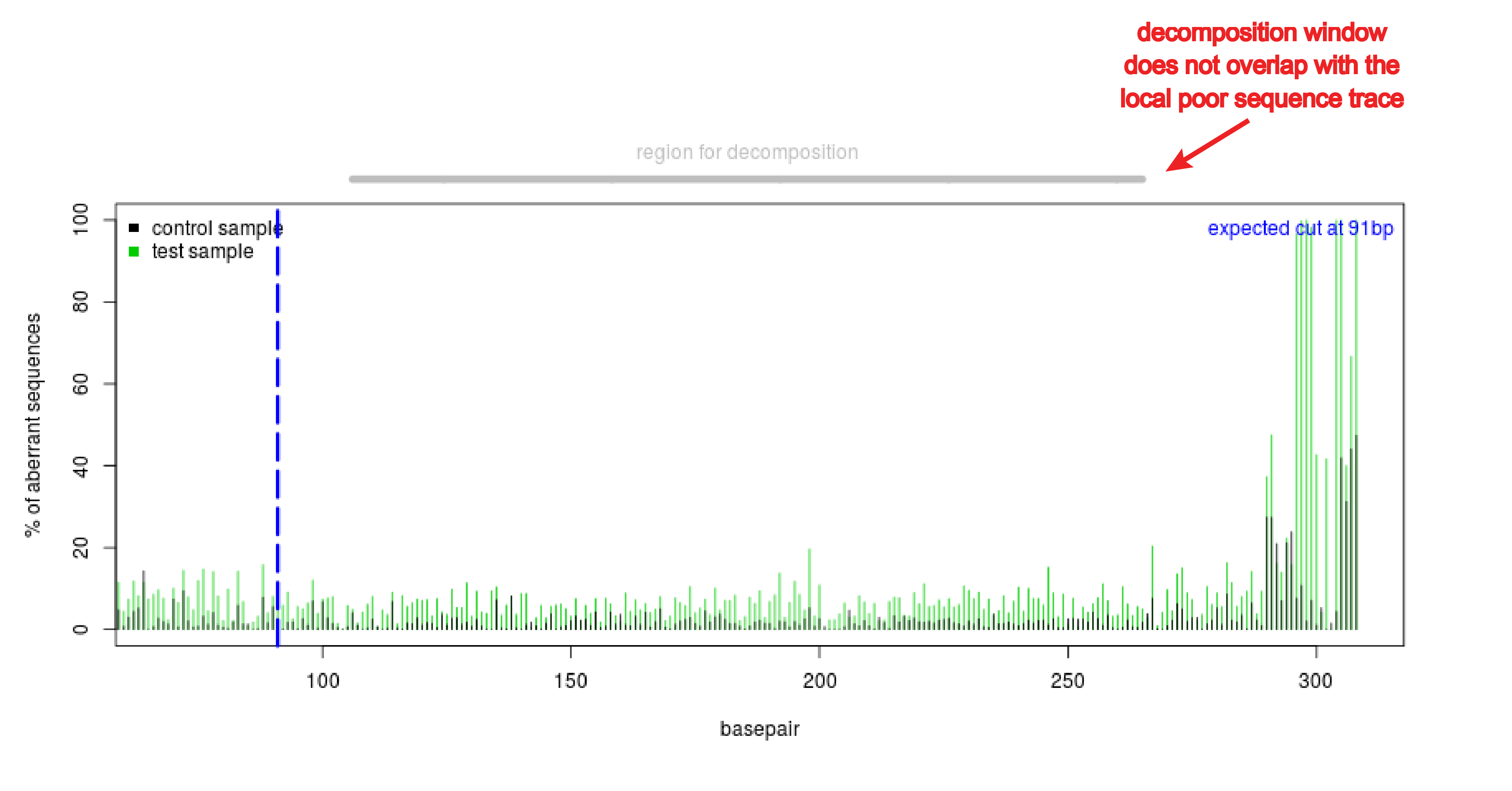
Analysis of gene modification conditions using TIDE software. The bar... | Download Scientific Diagram

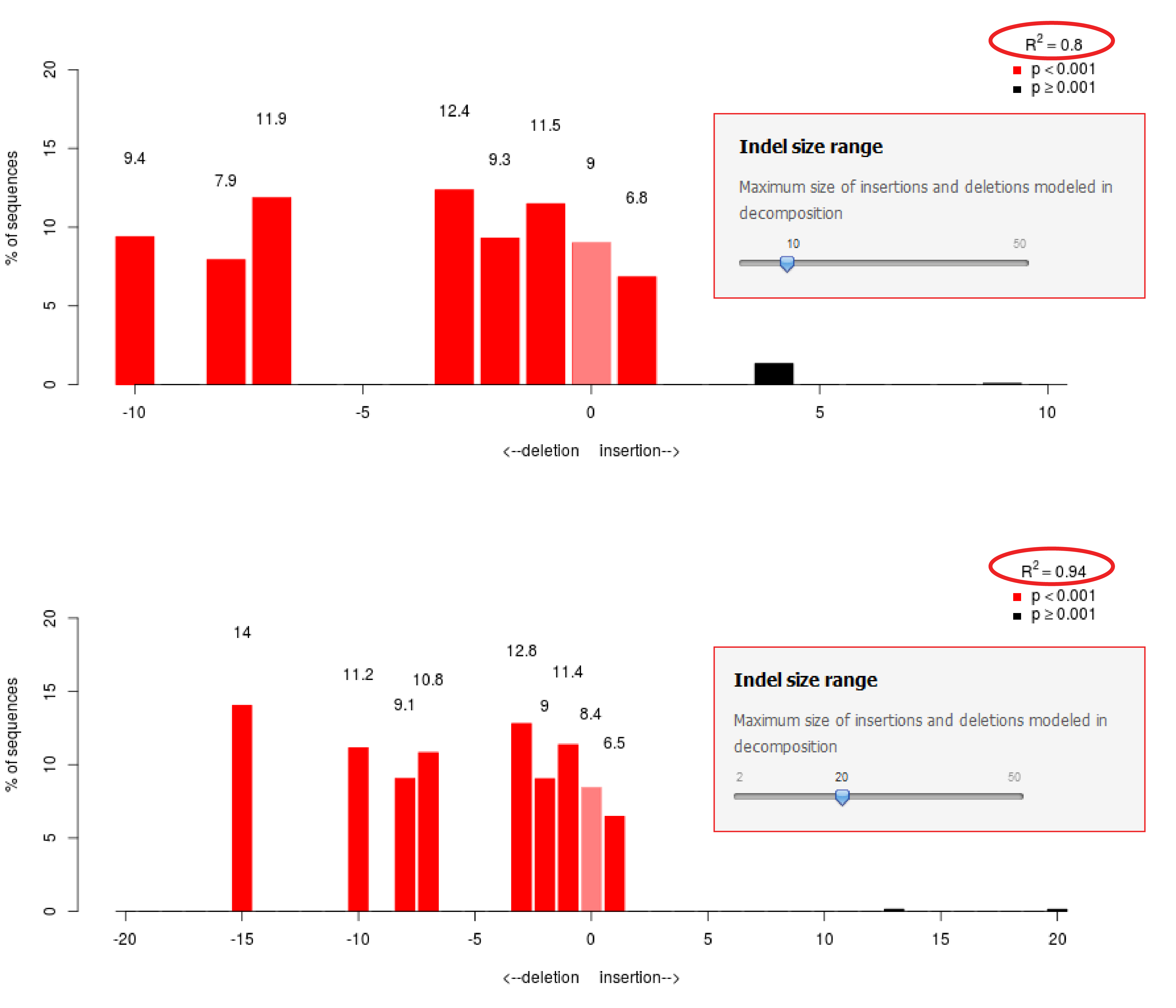


![A dual sgRNA approach for functional genomics in Arabidopsis thaliana [OPEN] | bioRxiv A dual sgRNA approach for functional genomics in Arabidopsis thaliana [OPEN] | bioRxiv](https://www.biorxiv.org/content/biorxiv/early/2017/08/21/172676/F1.large.jpg)